Launch a random search
Contents
# Uncomment this cell if running in Google Colab
!pip install clinicadl==1.6.1
Launch a random search#
The previous section focused on a way to debug non-automated architecture search. However, if you have enough computational power, you may want to launch an automated architecture search to save your time. This is the point of the random search method of ClinicaDL.
Warning
Non-optimal result: A random search may allow to find a better performing network, however there is no guarantee that this is the best performing network.
This notebook relies on the synthetic data generated in the previous notebook. If you did not run it, uncomment the following cell to generate the corresponding dataset. If you want to generate a bigger synthetic CAPS, please check this notebook
!curl -k https://aramislab.paris.inria.fr/clinicadl/files/handbook_2023/data_oasis/CAPS_extracted.tar.gz -o oasisCaps.tar.gz
!tar xf oasisCaps.tar.gz
!clinicadl generate trivial data_oasis/CAPS_example data/synthetic --n_subjects 4 --preprocessing t1-linear
Define the hyperparameter space#
A random search is performed according to hyperparameters of the network that are sampled from a pre-defined space. For example, you may want your random network to have maximum 3 fully-convolutional layers as you don’t have enough memory to tackle more.
This hyperparameter space is defined in a TOML file that must be written in
your random search directory: random_search.toml.
The following function generate_dict generates a dictionary that will be
used to random_search.toml for this tutorial. To accelerate the training
task we will use a single CNN on the default region of interest, the
hippocampi.
def generate_dict(gpu_avail, caps_dir, tsv_path, preprocessing_json):
return {
"Random_Search":{
"caps_directory": caps_dir,
"tsv_path": tsv_path,
"diagnoses": ['AD', 'CN'],
"preprocessing_json": preprocessing_json,
"network_task": "classification",
"n_convblocks": [1, 5], # Number of convolutional blocks
"first_conv_width": [8, 16, 32, 64], # Number of channels in the first convolutional block
"n_fcblocks": [1, 3]
}, # Number of (fully-connected + activation) layers
"Computational":{
"gpu": gpu_avail
},
"Optimization":{
"epochs": 30,
"learning_rate": [4, 6] # Threshold at which a region is selected if its corresponding balanced accuracy is higher.
},
"Cross_validation":{
"n_splits":3
}
}
In this default dictionary we set all the arguments that are mandatory for the random search. Hyperparameters for which a space is not defined will automatically have their default value in all cases.
Hyperparameters can be sampled in 4 different ways:
choice samples one element from a list (ex:
first_conv_width),uniform draws samples from a uniform distribution over the interval [min, max] (ex:
selection_threshold),exponent draws x from a uniform distribution over the interval [min, max] and return $10^{-x}$ (ex:
learning_rate),randint returns an integer in [min, max] (ex:
n_conv_blocks).
The values of some variables are also fixed, meaning that they cannot be sampled and that they should be given a unique value.
The values of the variables that can be set in random_search.toml correspond to the options of the train function. Values of the Computational resources section can also be redefined using the command line. Some variables were also added to sample the architecture of the network.
In the default dictionary, the learning rate will be sampled between $10^{-4}$ and $10^{-6}$.
This dictionary is written as a TOML file in the launch_dir of the
random-search.
You can define differently other hyperparameters by looking at the
documentation.
import os
import torch
# Check if a GPU is available
gpu_avail = torch.cuda.is_available()
mode = "image"
caps_dir = "data/synthetic"
tsv_path = "data/split/3_fold"
preprocessing_json = "extract_T1linear_image.json"
os.makedirs("random_search", exist_ok=True)
default_dict = generate_dict(gpu_avail, caps_dir, tsv_path, preprocessing_json)
# Add some changes here
import toml
toml_string = toml.dumps(default_dict) # Output to a string
output_file_name = "random_search/random_search.toml"
with open(output_file_name, "w") as toml_file:
toml.dump(default_dict, toml_file)
Prerequisites#
You need to execute the clinicadl tsvtool getlabels
and clinicadl tsvtool {split|kfold}
commands prior to running this task to have the correct TSV file organization.
Moreover, there should be a CAPS, obtained running the t1-linear pipeline of
ClinicaDL.
Running the task#
This task can be run with the following command line:
clinicadl random-search [OPTIONS] LAUNCH_DIRECTORY NAME
where:
launch_directory(Path) is the parent directory of output folder containing the filerandom_search.toml.name(str) is the name of the output folder containing the experiment.
Content of random_search.toml#
random_search.toml must be present in launch_dir before running the
command.
Mandatory variables:
network_task(str) is the task learnt by the network. Must be chosen betweenclassificationandregression(random sampling forreconstructionis not implemented yet). Sampling function:fixed.caps_directory(str) is the input folder containing the neuroimaging data in a CAPS hierarchy. Sampling function:fixed.preprocessing_json(str) corresponds to the JSON file produced byclinicadl extractused for the search. Sampling function:fixed.tsv_path(str) is the input folder of a TSV file tree generated byclinicadl tsvtool {split|kfold}. Sampling function:fixed.diagnoses(list of str) is the list of the labels that will be used for training. Sampling function:fixed.epochs(int) is the maximum number of epochs. Sampling function:fixed.n_convblocks(int) is the number of convolutional blocks in CNN. Sampling function:randint.first_conv_width(int) is the number of kernels in the first convolutional layer. Sampling function:choice.n_fcblocks(int) is the number of fully-connected layers at the end of the CNN. Sampling function:randint.
Train & evaluate a random network#
Based on the hyperparameter space described in random_search.json, you will
now be able to train a random network. To do so the following command can be
run:
!clinicadl random-search random_search maps_random_search
A new folder test has been created in launch_dir. As for any network
trained with ClinicaDL it is possible to evaluate its performance on a test
set:
# Evaluate the network performance on the 2 test images
!clinicadl predict random_search/maps_random_search test --participant_tsv data/split/test.tsv --caps_directory data/synthetic --selection_metrics "loss" --no-gpu
import pandas as pd
split = 0
predictions = pd.read_csv("./random_search/maps_random_search/split-%i/best-loss/test_image_level_prediction.tsv" % split, sep="\t")
display(predictions)
metrics = pd.read_csv("./random_search/maps_random_search/split-%i/best-loss/test_image_level_metrics.tsv" % split, sep="\t")
display(metrics)
Analysis of the random network#
The architecture of the network can be retrieved from the maps.json
file in the folder corresponding to a random job.
The architecture can be fully retrieved with 4 keys:
convolutionsis a dictionary describing each convolutional block,network_normalizationis the type of normalization layer used in convolutional blocks,n_fcblocksis the number of fully-connected layers,dropoutis the dropout rate applied at the dropout layer.
One convolutional block is described by the following values:
in_channelsis the number of channels of the input (if set to null corresponds to the number of channels of the input data),out_channelsis the number of channels in the output of the convolutional block. It corresponds to 2 *in_channelsexcept for the first channel chosen fromfirst_conv_width, and if it becomes greater thanchannels_limit.n_convcorresponds to the number of convolutions in the convolutional block,d_reductionis the dimension reduction applied in the block.
Convolutional block - example 1#
Convolutional block dictionary:
{
"in_channels": 16,
"out_channels": 32,
"n_conv": 2,
"d_reduction": "MaxPooling"
}
(network_normalization is set to InstanceNorm)
Corresponding architecture drawing:

Convolutional block - example 1#
Convolutional block dictionary:
{
"in_channels": 32,
"out_channels": 64,
"n_conv": 3,
"d_reduction": "stride"
}
(network_normalization is set to BatchNorm)
Corresponding architecture drawing:
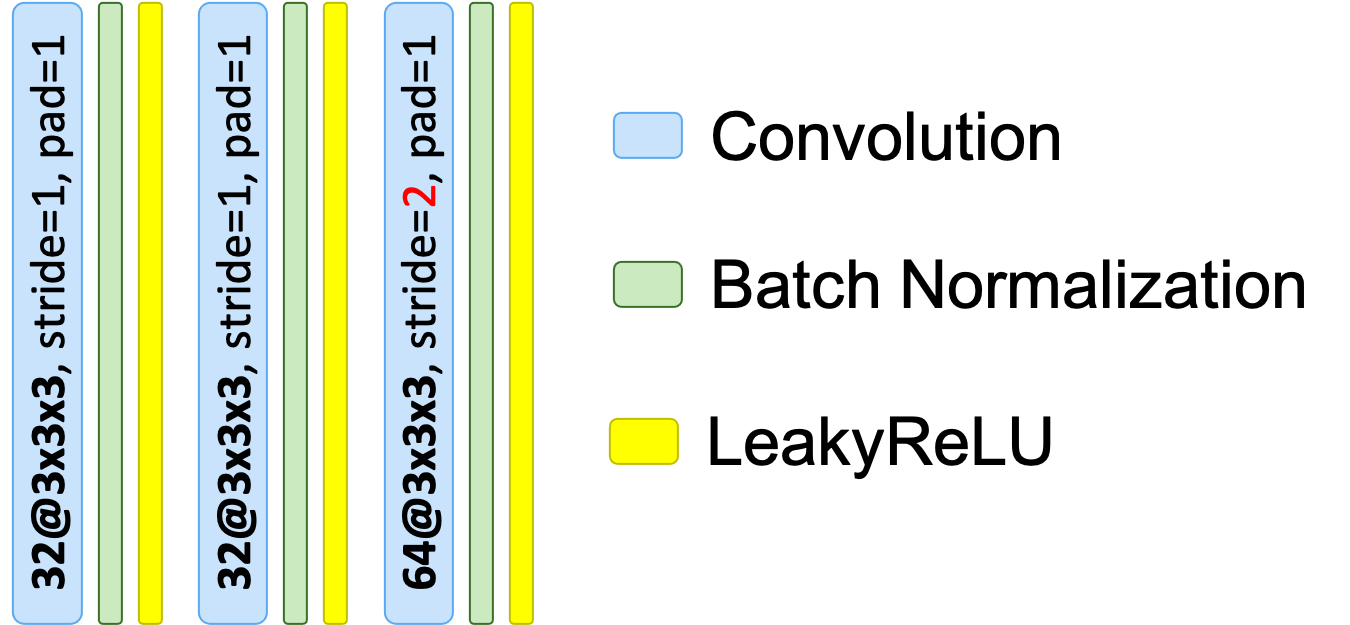
A simple way to better visualize your random architecture is to construct it
using create_model function from ClinicaDL. This function needs the list of
options of the model stored in the JSON file as well as the size of the input.
!pip install torchsummary
from clinicadl.utils.maps_manager.maps_manager_utils import read_json
from clinicadl.utils.caps_dataset.data import return_dataset, get_transforms
from torchsummary import summary
import argparse
import warnings
def create_model(options, initial_shape):
"""
Creates model object from the model_name.
:param options: (Namespace) arguments needed to create the model.
:param initial_shape: (array-like) shape of the input data.
:return: (Module) the model object
"""
from clinicadl.utils.network.cnn.random import RandomArchitecture
if not hasattr(options, "model"):
model = RandomArchitecture(options.convolutions, options.n_fcblocks, initial_shape,
options.dropout, options.network_normalization, n_classes=2)
else:
try:
model = eval(options.model)(dropout=options.dropout)
except NameError:
raise NotImplementedError(
'The model wanted %s has not been implemented.' % options.model)
if options.gpu:
model.cuda()
else:
model.cpu()
return model
warnings.filterwarnings('ignore')
# Read model options
options = argparse.Namespace()
model_options = read_json(options, json_path="random_search/test/commandline.json")
model_options.gpu = True
# Find data input size
_, transformations = get_transforms(mode, not model_options.unnormalize)
dataset = return_dataset(mode, caps_dir, tsv_path,
preprocessing_json, transformations, model_options)
input_size = dataset.size
# Create model and print summary
model = create_model(model_options, input_size)
summary(model, input_size)


