Generate a synthetic dataset
Contents
# Uncomment this cell if running in Google Colab
!pip install clinicadl==1.6.1
Generate a synthetic dataset#
Looking for new network architectures to improve performance on a
deep learning task implies testing different sets of hyperparameters. This
takes a lot of time and we often end up with networks that don’t
converge. To avoid this pitfall, it is often advised to simplify the problem:
focus on a subset of data or a task that is more tractable than
the one that is currently explored. This is the purpose of clinicadl generate which creates synthetic, tractable data from real data to
check that developed networks are working on this simple case before going
further.
With ClinicaDL, you can generate three types of synthetic data sets for a
binary classification task depending on the option chosen: trivial, random or
shepplogan.
If you ran the previous notebook, you must have a folder called
CAPS_example in the data_oasis directory (otherwise uncomment the next cell
to download a local version of the necessary folders).
!curl -k https://aramislab.paris.inria.fr/clinicadl/files/handbook_2023/data_oasis/CAPS_example.tar.gz -o oasisCaps.tar.gz
!tar xf oasisCaps.tar.gz
Generate trivial data#
Tractable data can be generated from real data with ClinicaDL. The command generates a synthetic dataset for a binary classification task from a CAPS-formatted dataset. It produces a new CAPS containing trivial data which should be perfectly classified. Each label corresponds to brain images whose intensities in the right or the left hemisphere were strongly decreased. Trivial data is useful for debugging a framework: hyper parameters can be more easily tested as fewer data samples are required and convergence should be reached faster as the classification task is simpler.
Warning
You need to execute the clinica run pipeline before running this task.
Moreover, the trivial option can synthesize at most n images per label,
where n is the total number of images in the input CAPS.
Running the task#
clinicadl generate trivial <caps_directory> <output_directory> --n_subjects <n_subjects>
where:
caps_directoryis the output folder containing the results ofclinica runin a CAPS hierarchy,output_directoryis the folder where the synthetic CAPS is stored,n_subjectsis the number of subjects per label in the synthetic dataset. Default value: 300.
Warning
n_subjects cannot be higher than the number of subjects in the initial
dataset. Indeed in each synthetic class, each synthetic image is derived
from a real image.
!clinicadl generate trivial data_oasis/CAPS_example data/synthetic --n_subjects 4 --preprocessing t1-linear
Reproduce the tsv file system necessary for training#
In order to train a network, meta data must be organized in a file system
generated by clinicadl tsvtools. For more information on the following
commands, please refer to the section Define your
population.
Get the labels AD and CN#
get-labels command needs a BIDS folder as an argument in order to create the
missing_mods directory and the merged_tsv file, but if you already
have these, you can give an empty folder as argument and provide the paths
to the required files separately as keyword arguments.
!mkdir data/fake_bids
!clinicadl tsvtools get-labels data/fake_bids data --missing_mods data/synthetic/missing_mods --merged_tsv data/synthetic/data.tsv --modality synthetic
# Split train and test data
!clinicadl tsvtools split data/labels.tsv --n_test 0.25 --subset_name test
# Split train and validation data in a 3-fold cross-validation
!clinicadl tsvtools kfold data/split/train.tsv --n_splits 3
Train a model on synthetic data#
Once data was generated and split, it is possible to train a model using
clinicadl train and evaluate its performance with clinicadl interpret. For
more information on the following command lines please read the sections
Classification with a CNN on 2D slice and
Regression with 3D images.
The following clinicadl train command uses a pre-build architecture of ClinicaDL Conv4_FC3.
You can also implement your own models by following the instructions of this
section.
First, we need to run prepare-data to extract the tensors from the images:
!clinicadl prepare-data image data/synthetic t1-linear --extract_json extract_T1linear_image
Then, we will train the network with the synthetic data. If you failed to generate a trivial dataset, please uncomment the next cell.
# !curl -k https://aramislab.paris.inria.fr/clinicadl/files/handbook_2023/data/synthetic.tar.gz -o synthetic.tar.gz
# !mkdir data
# !tar xf synthetic.tar.gz -C data
# !mkdir data/fake_bids
# !clinicadl tsvtools get-labels data/fake_bids data --missing_mods data/synthetic/missing_mods --merged_tsv data/synthetic/data.tsv --modality synthetic
# !clinicadl tsvtools split data/labels.tsv --n_test 0.25 --subset_name test
# !clinicadl tsvtools kfold data/split/train.tsv --n_splits 3
# # no need to run prepare-data
# Train a network with synthetic data (remove --no-gpu option if you do have access to a gpu)
!clinicadl train classification data/synthetic extract_T1linear_image data/split/3_fold data/synthetic_maps --architecture Conv4_FC3 --n_splits 3 --split 0 --no-gpu
As the number of images is very small (4 per class), we do not rely on the accuracy to select the model. Instead we evaluate the model which obtained the best loss.
# Evaluate the network performance on the 2 test images
!clinicadl predict data/synthetic_maps test --caps_directory ./data/synthetic --participants_tsv ./data/split/test_baseline.tsv --selection_metrics "loss"
import pandas as pd
fold = 0
predictions = pd.read_csv("./data/synthetic_maps/split-%i/best-loss/test/test_image_level_prediction.tsv" % fold, sep="\t")
display(predictions)
metrics = pd.read_csv("./data/synthetic_maps/split-%i/best-loss/test/test_image_level_metrics.tsv" % fold, sep="\t")
display(metrics)
Generate random data#
This command generates a synthetic dataset for a binary classification task from a CAPS-formatted dataset. It produces a new CAPS containing random data which cannot be correctly classified. All the images from this dataset comes from the same image to which random noise is added. Then the images are randomly distributed between the two labels

Warning
You need to execute the clinica run pipeline prior to running this task.
Moreover, the random option can synthesize as
many images as wanted with only one input image.
Running the task#
clinicadl generate random <caps_directory> <generated_caps_directory>
where:
caps_directoryis the output folder containing the results ofclinica runin a CAPS hierarchy,generated_caps_directoryis the folder where the synthetic CAPS is stored.
!clinicadl generate random data_oasis/CAPS_example data/CAPS_random --n_subjects 5 --preprocessing t1-linear
The command generates 3D images of same size as the input images formatted as
NIfTI files. Then the clinicadl prepare-data command must be run to use the
synthetic data with ClinicaDL. Results are stored in the same folder hierarchy
as the input folder.
Generate Shepp-Logan data#
This command is named after the Shepp-Logan phantom, a standard image to test image reconstruction algorithms. It creates three subtypes of 2D images distributed between two labels. These three subtypes can be separated according to the top (framed in blue) and bottom (framed in orange) regions:
subtype 0: Top and Bottom regions are of maximum size,
subtype 1: Top region has its maximum size but Bottom is atrophied,
subtype 2: Bottom region has its maximum size but Top is atrophied.
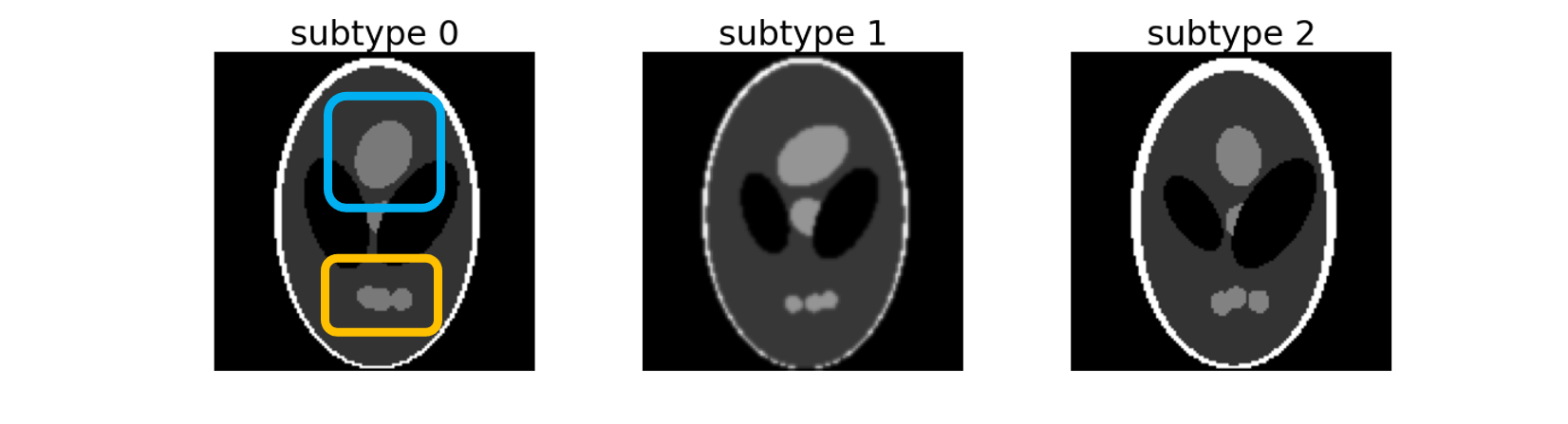
These three subtypes are spread between two labels which mimic the binary classification between Alzheimer’s disease patients (AD) with heterogeneous phenotypes and cognitively normal participants (CN). Default distributions are the following:
subtype |
0 |
1 |
2 |
|---|---|---|---|
AD |
5% |
85% |
10% |
CN |
100% |
0% |
0% |
The CN label is homogeneous, while the AD label is composed of a typical subtype (1), an atypical subtype (2) and normal looking images (0).
Running the task#
clinicadl generate shepplogan <generated_caps_directory>
where:
generated_caps_directoryis the folder where the synthetic CAPS is stored.
!clinicadl generate shepplogan data/CAPS_shepplogan --n_subjects 3


