# Uncomment this cell if running in Google Colab
# !pip install clinicadl==0.2.1
# !curl -k https://aramislab.paris.inria.fr/files/data/databases/tuto/dataOasis.tar.gz -o dataOasis.tar.gz
# !tar xf dataOasis.tar.gz
Launch a random search¶
The previous section focused on a way to debug non-automated architecture search. However, if you have enough computational power, you may want to launch an automated architecture search to save your time. This is the point of the random search method of clinicadl.
A random search may allow to find a better performing network, however there is no guarantee that this is the best performing network.
This notebook relies on the synthetic data generated in the previous notebook. If you did not run it, uncomment the following cell to generate the corresponding dataset.
import os
# os.makedirs("data", exist_ok=True)
# !curl -k https://aramislab.paris.inria.fr/files/data/databases/tuto/synthetic.tar.gz -o synthetic.tar.gz
# !tar xf synthetic.tar.gz -C data
Define the hyperparameter space¶
A random search is performed according to hyperparameters of the network that are sampled from a pre-defined space. For example, you may want your random network to have maximum 3 fully-convolutional layers as you don’t have enough memory to tackle more.
This hyperparameter space is defined in a JSON file that must be written in your random search directory: random_search.json.
The following function generate_dict generates a dictionnary that will be used to random_search.json for this tutorial. To accelerate the training task we will use a single CNN on the default region of interet, the hippocampi.
def generate_dict(mode, caps_dir, tsv_path, preprocessing):
return {
"caps_dir": caps_dir,
"tsv_path": tsv_path,
"diagnoses": ["AD", "CN"],
"preprocessing": preprocessing,
"mode": mode,
"network_type": "cnn",
"epochs": 30,
"learning_rate": [4, 6],
"n_convblocks": [1, 5], # Number of convolutional blocks
"first_conv_width": [8, 16, 32, 64], # Number of channels in the first convolutional block
"n_fcblocks": [1, 3], # Number of (fully-connected + activation) layers
"selection_threshold": [0.5, 1] # Threshold at which a region is selected if its corresponding
# balanced accuracy is higher.
}
In this default dictionnary we set all the arguments that are mandatory for the random search. Hyperparameters for which a space is not defined will automatically have their default value in all cases.
Hyperparameters can be sampled in 4 different ways:
choice samples one element from a list (ex:
first_conv_width),uniform draws samples from a uniform distribution over the interval [min, max] (ex:
selection_threshold),exponent draws x from a uniform distribution over the interval [min, max] and return \(10^{-x}\) (ex:
learning_rate),randint returns an integer in [min, max] (ex:
n_conv_blocks).
In the default dictionnary, the learning rate will be sampled between \(10^{-4}\) and \(10^{-6}\).
This dictionnary is written as a JSON file in the launch_dir of the random-search.
You can define differently other hyperparameters by looking at the documentation.
import os
import json
mode = "image"
caps_dir = "data/synthetic"
tsv_path = "data/synthetic/labels_list/train"
preprocessing = "t1-linear"
os.makedirs("random_search", exist_ok=True)
default_dict = generate_dict(mode, caps_dir, tsv_path, preprocessing)
# Add some changes here
json = json.dumps(default_dict, skipkeys=True, indent=4)
with open(os.path.join("random_search", "random_search.json"), "w") as f:
f.write(json)
Train & evaluate a random network¶
Based on the hyperparameter space described in random_search.json, you will now be able to train a random network. To do so the following command can be run:
clinicadl random-search <launch_dir> <name> --n_splits <n_splits>
where:
launch_diris the folder in which is locatedrandom_search.jsonand your future output jobs.output_directoryis the name of the folder of the job launched.n_splitsis the number of splits in the cross-validation procedure.
Other arguments, linked to computational resources can be specified when launching the random training.
!clinicadl random-search "random_search" "test" --n_splits 3 --split 0 -cpu -np 0 -v
A new folder test has been created in launch_dir. As for any network trained with ClinicaDL it is possible to evaluate its performance on a test set:
# Evaluate the network performance on the 2 test images
!clinicadl classify ./data/synthetic ./data/synthetic/labels_list/test ./random_search/test 'test' --selection_metrics "loss" -cpu
import pandas as pd
fold = 0
predictions = pd.read_csv("./random_search/test/fold-%i/cnn_classification/best_loss/test_image_level_prediction.tsv" % fold, sep="\t")
display(predictions)
metrics = pd.read_csv("./random_search/test/fold-%i/cnn_classification/best_loss/test_image_level_metrics.tsv" % fold, sep="\t")
display(metrics)
| participant_id | session_id | true_label | predicted_label | |
|---|---|---|---|---|
| 0 | sub-TRIV4 | ses-M00 | 1 | 1 |
| 1 | sub-TRIV5 | ses-M00 | 0 | 0 |
| accuracy | balanced_accuracy | sensitivity | specificity | ppv | npv | total_loss | total_kl_loss | image_id | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.012792 | 0 | NaN |
Analysis of the random network¶
The architecture of the network can be retrieved from the commandline.json file in the folder corresponding to a random job.
The architecture can be fully retrieved with 4 keys:
convolutionsis a dictionnary describing each convolutional block,network_normalizationis the type of normalization layer used in covolutional blocks,n_fcblocksis the number of fully-connected layers,dropoutis the dropout rate applied at the dropout layer.
One convolutional block is described by the following values:
in_channelsis the number of channels of the input (if set to null corresponds to the number of channels of the input data),out_channelsis the number of channels in the output of the convolutional block. It corresponds to 2 *in_channelsexcept for the first channel chosen fromfirst_conv_width, and if it becomes greater thanchannels_limit.n_convcorresponds to the number of convolutions in the convolutional block,d_reductionis the dimension reduction applied in the block.
Convolutional block - example 1¶
Convolutional block dictionnary:
{
"in_channels": 16,
"out_channels": 32,
"n_conv": 2,
"d_reduction": "MaxPooling"
}
(network_normalization is set to InstanceNorm)
Corresponding architecture drawing:

Convolutional block - example 1¶
Convolutional block dictionnary:
{
"in_channels": 32,
"out_channels": 64,
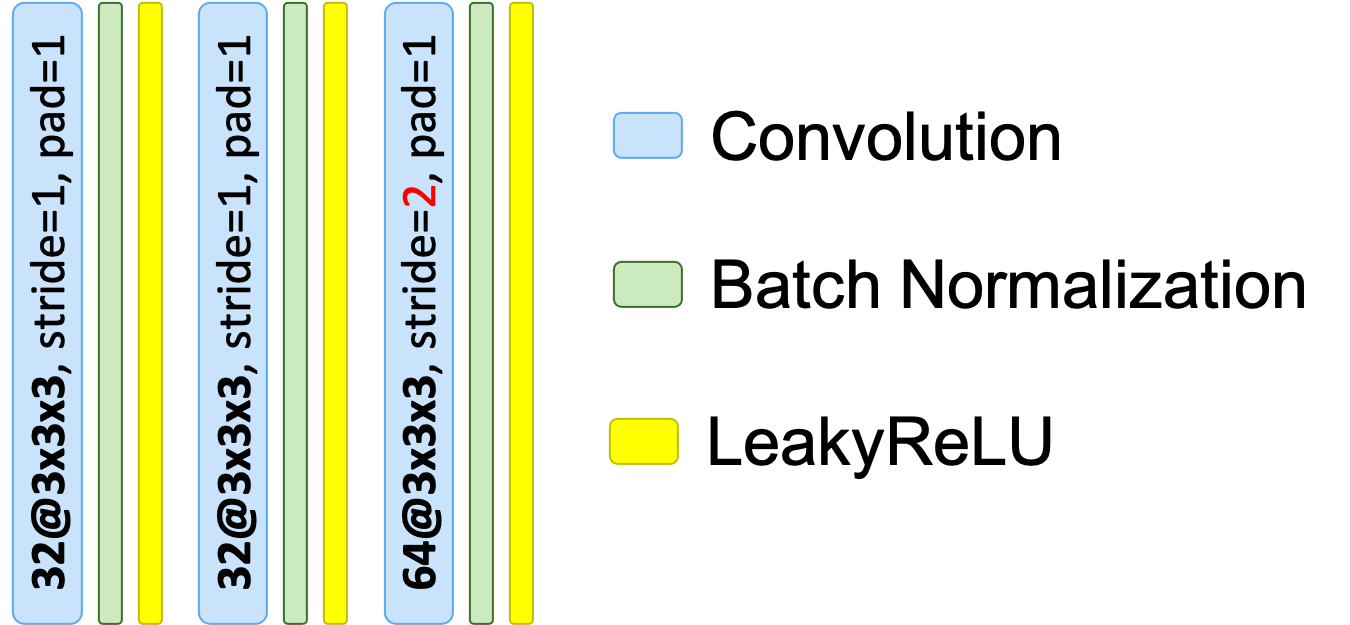
"n_conv": 3,
"d_reduction": "stride"
}
(network_normalization is set to BatchNorm)
Corresponding architecture drawing:
A simple way to better visualize your random architecture is to construct it using create_model function from ClinicaDL. This function needs the list of options of the model stored in the JSON file as well as the size of the input.
# !pip install torchsummary
from clinicadl.tools.deep_learning.iotools import read_json
from clinicadl.tools.deep_learning.models import create_model
from clinicadl.tools.deep_learning.data import return_dataset, get_transforms
from torchsummary import summary
import argparse
import warnings
warnings.filterwarnings('ignore')
# Read model options
options = argparse.Namespace()
model_options = read_json(options, json_path="random_search/test/commandline.json")
model_options.gpu = True
# Find data input size
_, transformations = get_transforms(mode, not model_options.unnormalize)
dataset = return_dataset(mode, caps_dir, os.path.join(tsv_path, "AD.tsv"),
preprocessing, transformations, model_options)
input_size = dataset.size
# Create model and print summary
model = create_model(model_options, input_size)
summary(model, input_size)
----------------------------------------------------------------
Layer (type) Output Shape Param #
================================================================
Conv3d-1 [-1, 32, 169, 208, 179] 896
BatchNorm3d-2 [-1, 32, 169, 208, 179] 64
LeakyReLU-3 [-1, 32, 169, 208, 179] 0
ConstantPad3d-4 [-1, 32, 170, 208, 180] 0
MaxPool3d-5 [-1, 32, 85, 104, 90] 0
PadMaxPool3d-6 [-1, 32, 85, 104, 90] 0
Conv3d-7 [-1, 64, 85, 104, 90] 55,360
BatchNorm3d-8 [-1, 64, 85, 104, 90] 128
LeakyReLU-9 [-1, 64, 85, 104, 90] 0
ConstantPad3d-10 [-1, 64, 86, 104, 90] 0
MaxPool3d-11 [-1, 64, 43, 52, 45] 0
PadMaxPool3d-12 [-1, 64, 43, 52, 45] 0
Conv3d-13 [-1, 128, 43, 52, 45] 221,312
BatchNorm3d-14 [-1, 128, 43, 52, 45] 256
LeakyReLU-15 [-1, 128, 43, 52, 45] 0
ConstantPad3d-16 [-1, 128, 44, 52, 46] 0
MaxPool3d-17 [-1, 128, 22, 26, 23] 0
PadMaxPool3d-18 [-1, 128, 22, 26, 23] 0
Conv3d-19 [-1, 256, 22, 26, 23] 884,992
BatchNorm3d-20 [-1, 256, 22, 26, 23] 512
LeakyReLU-21 [-1, 256, 22, 26, 23] 0
ConstantPad3d-22 [-1, 256, 22, 26, 24] 0
MaxPool3d-23 [-1, 256, 11, 13, 12] 0
PadMaxPool3d-24 [-1, 256, 11, 13, 12] 0
Conv3d-25 [-1, 512, 11, 13, 12] 3,539,456
BatchNorm3d-26 [-1, 512, 11, 13, 12] 1,024
LeakyReLU-27 [-1, 512, 11, 13, 12] 0
ConstantPad3d-28 [-1, 512, 12, 14, 12] 0
MaxPool3d-29 [-1, 512, 6, 7, 6] 0
PadMaxPool3d-30 [-1, 512, 6, 7, 6] 0
Flatten-31 [-1, 129024] 0
Dropout-32 [-1, 129024] 0
Linear-33 [-1, 3217] 415,073,425
LeakyReLU-34 [-1, 3217] 0
Linear-35 [-1, 80] 257,440
LeakyReLU-36 [-1, 80] 0
Linear-37 [-1, 2] 162
================================================================
Total params: 420,035,027
Trainable params: 420,035,027
Non-trainable params: 0
----------------------------------------------------------------
Input size (MB): 24.00
Forward/backward pass size (MB): 8773.51
Params size (MB): 1602.31
Estimated Total Size (MB): 10399.82
----------------------------------------------------------------
