# Uncomment the next lines if running in Google Colab
# !pip install clinicadl==0.2.1
# !curl -k https://aramislab.paris.inria.fr/files/data/databases/tuto/dataOasis.tar.gz -o dataOasis.tar.gz
# !tar xf dataOasis.tar.gz
Define your population¶
This notebook is an introduction to tools that can be used to identify relevant samples and split them between training, validation and test cohorts. This step is mandatory preliminary to training to avoid issues such as lack of clinical meaning or data leakage.
In the following, we will see how to
extract samples corresponding to labels of interest from a BIDS hierarchy,
split these samples between training, validation and test sets
using tools available in clinica and clinicadl.
Merge metadata from a BIDS hierarchy with clinica iotools¶
In a BIDS hierarchy, demographic, clinical and imaging metadata are stored in TSV files located at different levels of the hierarchy depending on whether they are specific to a subject (e.g. gender), a session (e.g. diagnosis) or a scan (e.g. acquisitions parameters).
The following command line can be used to merge all the metadata in a single TSV file:
clinica iotools merge-tsv <bids_directory> <output_tsv>
where:
bids_directoryis the input folder containing the dataset in a BIDS hierarchy.output_tsvis the path of the output tsv. If a directory is specified instead of a file name, the default name for the file created will bemerge-tsv.tsv.
In the preprocessing section an example BIDS of 4 subjects from OASIS-1 was generated (if you did not run interactively that section, download the dataset by uncomment the next cell):
# !curl -k https://aramislab.paris.inria.fr/files/data/databases/tuto/OasisBids.tar.gz -o OasisBids.tar.gz
# !tar xf OasisBids.tar.gz
Execute the following command to gather metadata included in this BIDS:
# Merge meta-data information
!clinica iotools merge-tsv OasisBids_example example_merged.tsv
We want to restrict the list of the sessions used to those including a T1-MR image. Then the following command is needed to identify which modalities are present for each session:
clinica iotools check-missing-modalities <bids_directory> <output_directory>
where:
bids_directoryis the input folder of a BIDS compliant dataset.output_directoryis the output folder.
Execute the following command to find which sessions include a T1-MR image on the example BIDS of OASIS:
# Find missing modalities
!clinica iotools check-missing-modalities OasisBids_example example_missing_mods
The output of this command, data/example_missing_mods, is a folder in which a series of tsv files is written (one file per session label containing one row per subject and one column per modality).
Get labels with clinicadl tsvtool on OASIS¶
In this section we will now get the labels from the whole OASIS dataset on which clinica iotools merge-tsv and clinica iotools check-missing-modalities were already performed.
The outputs of the corresponding pipelines can be found respectively in data/OASIS_BIDS.tsv and data/OASIS_missing_mods, and are provided on GitHub with the notebooks.
Restrict the OASIS dataset to older adults¶
OASIS-1 consists of 416 subjects aged 18 to 96:
subjects |
age |
sex |
MMSE |
CDR |
|
|---|---|---|---|---|---|
AD |
100 |
76.8 ± 7.1 [62.0, 96.0] |
59F / 41M |
24.3 ± 4.1 [14, 30] |
0: 0, 0.5: 70, 1: 28, 2: 2 |
CN |
316 |
45.1 ± 23.9 [18.0, 94.0] |
197F / 119M |
29.6 ± 0.9 [25, 30] |
0: 316 |
As you can see, CN participants are on average younger than AD participants, which makes sense as AD mainly affects older adults. However, this is an issue as aging also causes brain atrophy. The classifier may mix the signal due to healthy brain aging and Alzheimer’s disease on such dataset, which could lead to an over-estimation of the performance.
To avoid this bias, CN participants younger than the youngest AD patient were removed. This restriction can be run with the following command line:
clinicadl tsvtool restrict <dataset> <merged_tsv> <results_path>
where:
dataset(str) is the name of the dataset. Choices areOASISorAIBL.merged_tsv(str) is the output file of theclinica iotools merge-tsvcommand.results_path(str) is the path to the output tsv file (filename included). This tsv file comprises the same columns asmerged_tsv.
Execute the following cell to apply the restriction to OASIS-1:
!clinicadl tsvtool restrict OASIS data/OASIS_BIDS.tsv data/OASIS_restricted_BIDS.tsv
Some other sessions were also excluded because the preprocessing operations failed. The list of the images that were kept after the preprocessing is stored in data/OASIS_qc_output.tsv.
Get the labels¶
The 5 labels described in the first part of the course can be extracted with clinicadl using the command:
clinicadl tsvtool getlabels merged_tsv missing_mods results_path
where:
merged_tsvis the output file of theclinica iotools merge-tsvorclinicadl tsvtool restrictcommands.missing_modsis the folder containing the outputs of theclinica iotools missing-modscommand.results_pathis the path to the folder where output tsv files will be written.
Tip
You can increase the verbosity of the command by adding -v flag(s).
By default the pipeline only extracts the AD and CN labels, which corresponds to the only available labels in OASIS. Run the following cell to extract them in a new folder labels_lists from the restricted version of OASIS:
!clinicadl tsvtool getlabels data/OASIS_restricted_BIDS.tsv data/OASIS_missing_mods data/labels_lists --restriction_path data/OASIS_after_qc.tsv
For each diagnostic label, a file has been created comprising all the sessions that can be included in the classification task.
At the end of the command line another restriction was given to extract the labels only from sessions in data/OASIS_after_qc.tsv. This tsv file corresponds to the output of the quality check procedure that was manually cut to only keep the sessions passing the quality check. It depends on the preprocessing: here it concerns a run of t1-linear.
Analyze the population¶
The age bias in OASIS is well known and this is why the youngest CN participants were previously excluded. However, other biases may exist, especially after the quality check of the preprocessing which removed sessions from the dataset. Thus it is crucial to check before going further if there are other biases in the dataset.
clinicadl implements a tool to perform a demographic and clinical analysis of the population:
clinicadl tsvtool analysis <merged_tsv> <formatted_data_path> <results_path>
where:
merged_tsvis the output file of theclinica iotools merge-tsvorclinicadl tsvtool restrictcommands.formatted_data_pathis a folder containing one tsv file per label (output ofclinicadl tsvtool getlabels|split|kfold).results_pathis the path to the tsv file that will be written (filename included).
The following command will extract statistical values on the populations for each diagnostic label. Based on those it is possible to check that the dataset is suitable for the classification task.
# Run the analysis
!clinicadl tsvtool analysis data/OASIS_BIDS.tsv data/labels_lists data/OASIS_analysis.tsv
def display_table(table_path):
"""Custom function to display the clinicadl tsvtool analysis output"""
import pandas as pd
OASIS_analysis_df = pd.read_csv(table_path, sep='\t')
OASIS_analysis_df.set_index("diagnosis", drop=True, inplace=True)
columns = ["n_subjects", "n_scans",
"mean_age", "std_age", "min_age", "max_age",
"sexF", "sexM",
"mean_MMSE", "std_MMSE", "min_MMSE", "max_MMSE",
"CDR_0", "CDR_0.5", "CDR_1", "CDR_2", "CDR_3"]
# Print formatted table
format_columns = ["subjects", "scans", "age", "sex", "MMSE", "CDR"]
format_df = pd.DataFrame(index=OASIS_analysis_df.index, columns=format_columns)
for idx in OASIS_analysis_df.index.values:
row_str = "%i; %i; %.1f ± %.1f [%.1f, %.1f]; %iF / %iM; %.1f ± %.1f [%.1f, %.1f]; 0: %i, 0.5: %i, 1: %i, 2:%i, 3:%i" % tuple([OASIS_analysis_df.loc[idx, col] for col in columns])
row_list = row_str.split(';')
format_df.loc[idx] = row_list
format_df.index.name = None
display(format_df)
display_table("data/OASIS_analysis.tsv")
| subjects | scans | age | sex | MMSE | CDR | |
|---|---|---|---|---|---|---|
| AD | 78 | 78 | 75.6 ± 7.0 [62.0, 96.0] | 43F / 35M | 24.4 ± 4.3 [14.0, 30.0] | 0: 0, 0.5: 56, 1: 20, 2:2, 3:0 |
| CN | 76 | 76 | 76.5 ± 8.4 [62.0, 94.0] | 62F / 14M | 29.0 ± 1.2 [25.0, 30.0] | 0: 76, 0.5: 0, 1: 0, 2:0, 3:0 |
There is no significant bias on age anymore, but do you notice any other problems?
There is still a difference in sex distribution and the network could learn a bias on sex such as "women are cognitively normal" and "men are demented". However, there are too few images in OASIS to continue removing sessions to equilibrate the groups.
To check that such bias is not learnt, it is possible to run a logistic regression after training between sex and the predicted label to check if they are correlated.
Split the data samples into training, validation and test sets¶
Now that the labels have been extracted and possible biases have been identified, data have to be split in different sets. This step is essential to guarantee the independence of the final evaluation.
In this notebook, data samples are divided between train, validation and test sets:
- The train set is used to update the weights,
- The validation set is used to stop the training process and select the best model,
- The test set is used after the end of the training process to perform an unbiased evaluation of the performance.
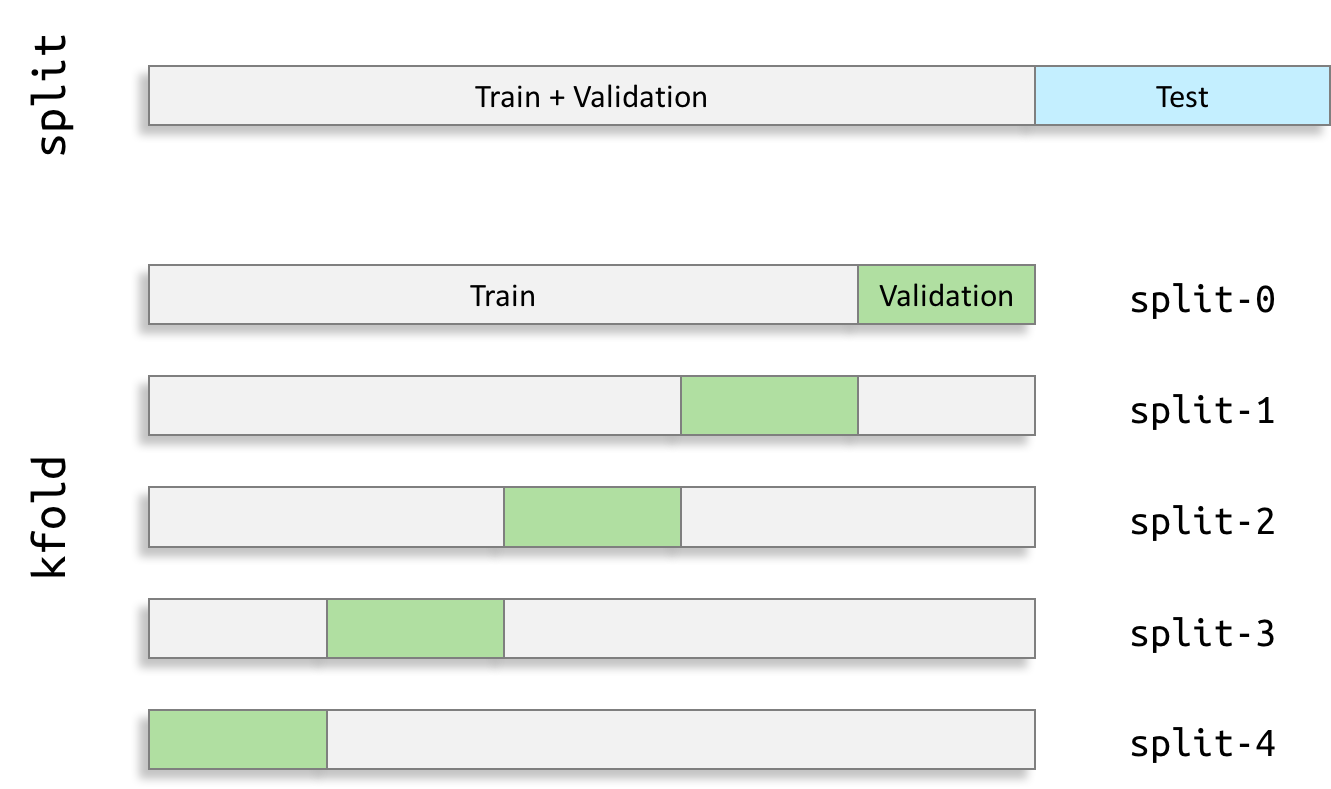
Due to the k-fold validation procedure, k trainings are conducted according to the k training/validation pairs generated. This leads to k different models that are evaluated on the test set at the end. The final test performance is then the mean value of these k models.
Tools that have been developed for this part are based on the guidelines of (Varoquaux et al., 2017).
Build the test set¶
The test set is obtained by performing a single split obtained with clinicadl tsvtool split:
clinicadl tsvtool split <merged_tsv> <formatted_data_path>
where:
merged_tsvis the output file of theclinica iotools merge-tsvorclinicadl tsvtool restrictcommands.formatted_data_pathis a folder containing one tsv file per label (output ofclinicadl tsvtool getlabels|split|kfold).
Each diagnostic label is split independently. Random splits are generated until there are non-significant differences between age and sex distributions between the test set and the train + validation set. Then three TSV files are written per label:
the baseline sessions of the test set,
the baseline sessions of the train + validation set,
the longitudinal sessions of the train + validation set.
In OASIS there is no longitudinal follow-up, hence the last two TSV files are identical.
Let’s create a test set including 20 subjects per label:
!clinicadl tsvtool split data/labels_lists --n_test 20
The differences between populations of the train + validation and test sets can be assessed to check that there is no discrepancies between the two sets.
!clinicadl tsvtool analysis data/OASIS_BIDS.tsv data/labels_lists/train data/OASIS_trainval_analysis.tsv
!clinicadl tsvtool analysis data/OASIS_BIDS.tsv data/labels_lists/test data/OASIS_test_analysis.tsv
print("Train + validation set")
display_table("data/OASIS_trainval_analysis.tsv")
print("Test set")
display_table("data/OASIS_test_analysis.tsv")
Train + validation set
| subjects | scans | age | sex | MMSE | CDR | |
|---|---|---|---|---|---|---|
| AD | 58 | 58 | 75.5 ± 7.1 [62.0, 96.0] | 31F / 27M | 24.3 ± 4.3 [14.0, 30.0] | 0: 0, 0.5: 41, 1: 16, 2:1, 3:0 |
| CN | 56 | 56 | 76.4 ± 8.4 [62.0, 94.0] | 46F / 10M | 29.0 ± 1.2 [25.0, 30.0] | 0: 56, 0.5: 0, 1: 0, 2:0, 3:0 |
Test set
| subjects | scans | age | sex | MMSE | CDR | |
|---|---|---|---|---|---|---|
| AD | 20 | 20 | 76.0 ± 6.8 [64.0, 90.0] | 12F / 8M | 24.6 ± 4.3 [15.0, 30.0] | 0: 0, 0.5: 15, 1: 4, 2:1, 3:0 |
| CN | 20 | 20 | 76.8 ± 8.4 [64.0, 90.0] | 16F / 4M | 28.8 ± 1.3 [26.0, 30.0] | 0: 20, 0.5: 0, 1: 0, 2:0, 3:0 |
If you are not satisfied with these populations, you can relaunch the test or change the parameters used to evaluate the difference between the distributions: p_val_threshold and t_val_threshold.
Only one test set was created in (Wen et al., 2020) to evaluate the final performance of one model. This is because architecture search was performed on the training + validation sets. As this operation is very costly and/or is done mostly manually, it was not possible to do it several times.
Build the validation sets¶
To better estimate the performance of the network, it is trained 5 times using a 5-fold cross-validation procedure. In this procedure, each sample is used once to validate and the other times to train the network. In the same way as for the single split, the TSV files can be processed with clinicadl:
clinicadl tsvtool kfold <formatted_data_path>
where formatted_data_path is a folder containing one tsv file per label (output of clinicadl tsvtool getlabels|split|kfold).
In a similar way than for the test split, three tsv files are written per label per split for each set:
the baseline sessions of the validation set,
the baseline sessions of the train set,
the longitudinal sessions of the train set.
Contrary to the test split, there is no attempt to control the similarity between the age and sex distributions. Indeed here we consider that averaging across the results of the 5 folds already reduces bias compared to a single data split.
!clinicadl tsvtool kfold data/labels_lists/train --n_splits 5
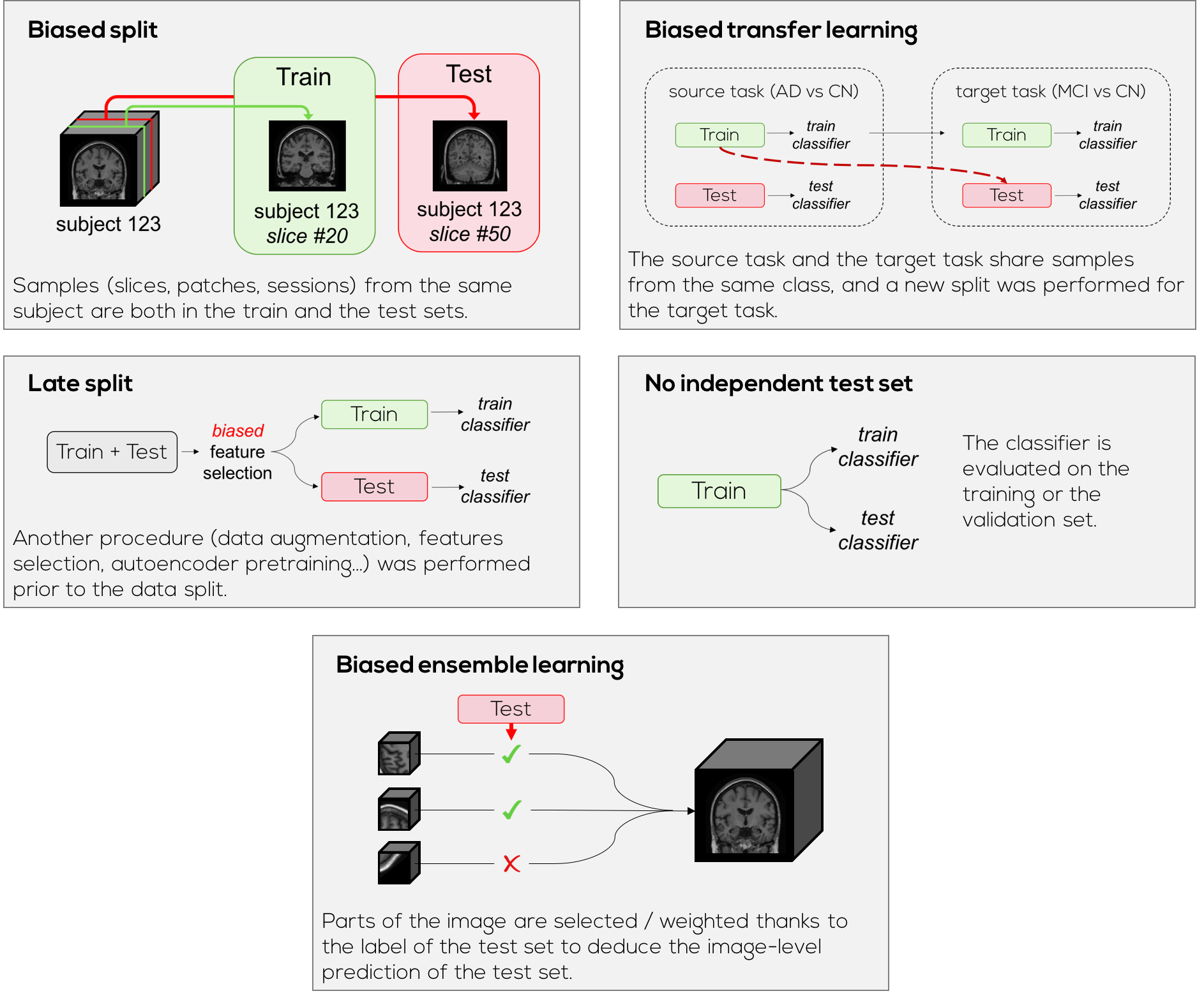
Check the absence of data leakage¶
In OASIS-1 there is no risk of data leakage due to the data split itself as there is only one session per subject. Also there is no MCI patients, hence there is no risk of data leakage during a transfer learning between a source task involving the MCI set and a target task involving at least one of its subsets (sMCI or pMCI). However for other datasets, it might be useful to check that there is no correlated data spread between the train and test sets.
A script in clinicadl has been created to check that there was no data leakage after the split steps. More specifically it checks that:
baseline datasets contain only one scan per subject,
no intersection exists between train and test sets,
MCI train subjects are absent from test sets of subcategories of MCI.
As it is not a common function, it has not been integrated to the general commandline. The next cell executes it on the splits generated in the previous sections.
from clinicadl.tools.tsv.test import run_test_suite
# Run check for train+val / test split
run_test_suite("./data/labels_lists", n_splits=0, subset_name="test")
# Run check for train / validation splits
run_test_suite("./data/labels_lists/train", n_splits=5, subset_name="validation")
If no Error was raised then none of the three conditions was broken. It is now possible to use the train and the validation sets to perform a classification task, and then to evaluate correctly the performance of the classifier on the test set.
Many procedures can cause data leakage and thus bias the performance, leading to impossible claims. It is crucial to check that the test set has not been contaminated by data that is correlated to the train and/or validation sets. You will find below examples of procedures that can lead to data leakage.