# Uncomment this cell if running in Google Colab
# !pip install clinicadl==0.2.1
# !curl -k https://aramislab.paris.inria.fr/files/data/databases/tuto/dataOasis.tar.gz -o dataOasis.tar.gz
# !tar xf dataOasis.tar.gz
Debug architecture search¶
Previous sections were focusing on pre-built architectures available in ClinicaDL. These architectures were trained and validated on ADNI, and gave similar test results on ADNI, AIBL and OASIS. However, they might not be transferrable to other problems on other datasets using other modalities, and this is why may want to search for new architectures and hyperparameters.
Looking for a new set of hyperparameters often means taking a lot of time training networks that are not converging. To avoid this pitfall, it is often advise to simplify the problem: focus on a subset of data / classification task that is more tractable than the one that is currently explored. This is the purpose of clinicadl generate which creates a set of synthetic, tractable data from real data to check that developed networks are working on this simple case before going further.
In this notebook, we call tractable data a set of pairs of images and labels that can be easily classified. In ClinicaDL, tractable data is generated from real brain images and consist in creating two classes in which the intensitites of the left or the right part of the brain are decreased.
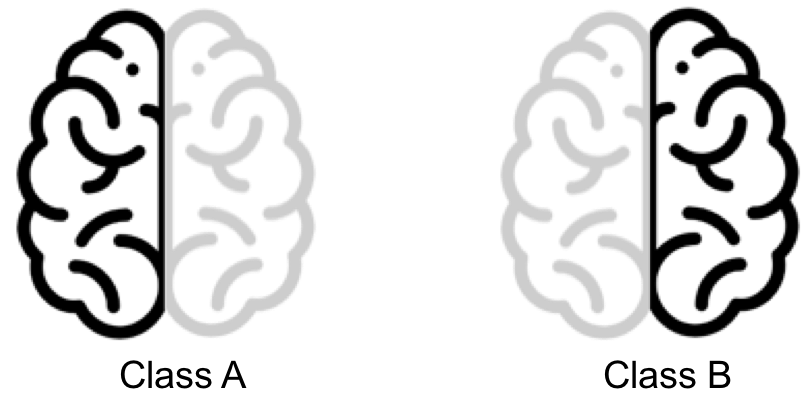
If you ran the previous notebook, you must have a folder called OasisCaps_example in the current directory (Otherwise uncomment the next cell to download a local version of the necessary folders).
# !curl -k https://aramislab.paris.inria.fr/files/data/databases/tuto/OasisCaps2.tar.gz -o OasisCaps2.tar.gz
# !tar xf OasisCaps2.tar.gz
Generate tractable data¶
Tractable data can be generated from real data with the following command line
clinicadl generate trivial <caps_directory> <output_directory> --n_subjects <n_subjects>
where:
caps_directoryis the output folder containing the results in a CAPS hierarchy.output_directoryis the folder where the synthetic CAPS is stored.n_subjectsis the number of subjects per label in the synthetic dataset. Default value: 300.
Warning
n_subjects cannot be higher than the number of subjects in the initial dataset. Indeed in each synthetic class, a synthetic image derives from a real image.
!clinicadl generate trivial OasisCaps_example t1-linear data/synthetic --n_subjects 4
This command will generate synthetic data in nifti files. To train the network, tensor files must be extracted with clinicadl extract. For more information on the following command line, please read the section Prepare your neuroimaging data.
This notebook will only focus on image-level classification. It is also possible to perform patch-level, slice-level and region-based classification. For more information read the corresponding section Train your own models.
!clinica run deeplearning-prepare-data data/synthetic t1-linear image --n_procs 2
Reproduce the tsv file system necessary to train¶
In order to train a network, meta data must be organized in a file system generated by clinicadl tsvtool. For more information on the following commands, please read the section Define your population.
# Get the labels AD and CN in separate files
!clinicadl tsvtool getlabels data/synthetic/data.tsv data/synthetic/missing_mods data/synthetic/labels_list --modality synthetic
# Split train and test data
!clinicadl tsvtool split data/synthetic/labels_list --n_test 0.25
# Split train and validation data in a 5-fold cross-validation
!clinicadl tsvtool kfold data/synthetic/labels_list/train --n_splits 3
Train a model on synthetic data¶
Once data was generated and split it is possible to train a model using clinicadl train and evaluate its performance with clinicadl classify. For more information on the following command lines please read the sections Train your own models and Perfom classification using pretrained models.
The following command uses a pre-build architecture of ClinicaDL Conv4_FC3. You can also implement your own models by following the instructions of this section.
# Train a network with synthetic data
!clinicadl train roi cnn data/synthetic t1-linear data/synthetic/labels_list/train results/synthetic Conv4_FC3 --n_splits 3 --split 0 -v
As the number of images is very small (4 per class), we do not rely on the accuracy to select the model. Instead we evaluate the model which obtained the best loss.
# Evaluate the network performance on the 2 test images
!clinicadl classify ./data/synthetic ./data/synthetic/labels_list/test ./results/synthetic 'test' --selection_metrics "loss"
import pandas as pd
fold = 0
predictions = pd.read_csv("./results/synthetic/fold-%i/cnn_classification/best_loss/test_image_level_prediction.tsv" % fold, sep="\t")
display(predictions)
metrics = pd.read_csv("./results/synthetic/fold-%i/cnn_classification/best_loss/test_image_level_metrics.tsv" % fold, sep="\t")
display(metrics)
| participant_id | session_id | true_label | predicted_label | |
|---|---|---|---|---|
| 0 | sub-TRIV4 | ses-M00 | 1 | 1 |
| 1 | sub-TRIV5 | ses-M00 | 0 | 0 |
| accuracy | balanced_accuracy | sensitivity | specificity | ppv | npv | |
|---|---|---|---|---|---|---|
| 0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
